Department of Synaptic Plasticity
Erin Schuman
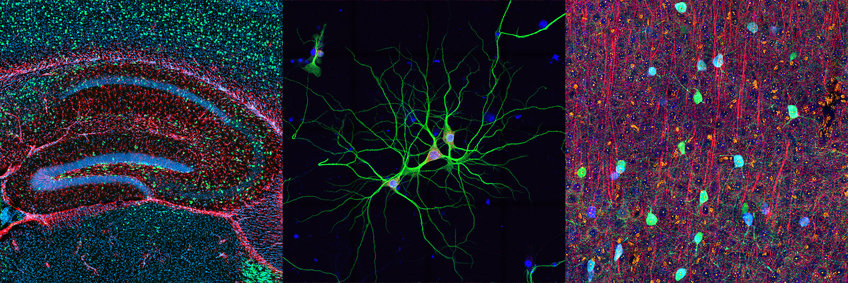
Erin Schuman lab’s long-standing research interest is the study of cellular mechanisms and neural circuits that underlie information processing and storage. The lab focuses on the molecular and cell biological processes that control protein synthesis and degradation in neurons and their synapses. The complex morphology of neurons, with most synapses located hundreds of microns from the cell body, presents a logistical challenge for the establishment, maintenance and modification of local synaptic proteomes. Neurons have solved this problem by localizing important cell biological machines, including ribosomes and proteasomes, within dendrites and axons.
Following on the lab’s initial discovery in 1996 that proteins made locally in dendrites are required for synaptic plasticity, they have pursued the identification of the mRNA and ribosome population present in neuronal dendrites and axons. The lab has discovered thousands of mRNAs present in dendrites and axons and characterized the unique regulatory elements present in these mRNAs.
In addition, they are elucidating the population of mRNAs translated in subcellular compartments as well as the nature and format of ribosomes present. In order to address the above questions, they have developed platforms to label, purify, identify and visualize newly synthesized proteins in neurons and other cells using non-canonical amino acid metabolic labelling, click chemistry, and mutation of cell-biological enzymes (the BONCAT and FUNCAT techniques). The lab’s current focus is on the nature and specialization of mRNA, protein synthesis and protein degradation machines and mechanisms in neurons. The lab also uses zebrafish as a system to study the molecular and cellular underpinnings of social behavior.
- Diversity of transcriptomes in dendrites, axons and synapses
- Diversity of proteomes in dendrites, axons and synapses
- Nature of neuronal ribosomes and translational machines
- Impact of transcriptome and proteome diversity on structure and function
- Protein degradation machines and the feedback mechanisms with protein synthesis
- Molecular and cellular underpinnings of social behavior
Selected recent papers (see here for all publications)
DOI Perez, J.D., tom Dieck, S., Alvarez Castelao, B., Tushev, G., Chan, I.C.W., and Schuman, E.M. Subcellular sequencing of single neurons reveals the dendritic transcriptome of GABAergic interneurons. eLife (2021).
DOI Anneser, L., Alcantara, I.C., Gemmer, A., Mirkes, K., Ryu, S., and Schuman, E.M. The neuropeptide Pth2 dynamically senses others via mechanosensation. Nature, 1-17. (2020).
DOI Alvarez-Castelao, B., tom Dieck, S., Fusco, C.M., Donlin-Asp, P.G., Perez, J.D., and Schuman, E.M. The switch-like expression of Heme-regulated kinase 1 mediates neuronal proteostasis following proteasome inhibition. eLife 2020;9:e52714 (2020).
DOI Biever, A., Glock, C., Tushev, G., Ciirdaeva, E., Dalmay, T., Langer, J.T. and Schuman, E.M. Monosomes actively translate synaptic mRNAs in neuronal processes. Science, 367 (6477), eaay 4991 (2020).
DOI Dörrbaum, A.R., Alvarez-Castelao, B., Nassim-Assir, B., Langer, J.D., and Schuman, E.M. Proteome dynamics during homeostatic scaling in cultured neurons. eLife 2020;9:e52939 (2020).
DOI Holt, C.E., Martin, K.C., and Schuman, E.M. Local translation in neurons: visualization and function. Nature Structural & Molecular Biology, 26: 557-566 (2019).
DOI Hafner, A.S., Donlin-Asp, P.G., Leitch, B., Herzog, E., and Schuman, E.M. Local protein synthesis is a ubiquitous feature of neuronal pre- and postsynaptic compartments. Science, 364, (6441), 650. (2019).
DOI Heumüller, M., Glock, C., Biever, A., Rangaraju, V. and Schuman, E.M. A genetically encodable protein synthesis inhibitor. Nature Methods,16, 699-702. (2019).
DOI Rangaraju, V., Lauterbach, M., and Schuman, E.M. Spatially stable mitochondrial compartments fuel local translation during synaptic plasticity. Cell, 176, 73-84. (2019).
DOI Alvarez-Castelao, B., Schanzenbaecher, C.T., Langer, J.D., and Schuman, E.M. Cell-type-specific metabolic labeling, detection and identification of nascent proteomes in vivo. Nature Protocols, 14(2), 556-575. (2019).
DOI Dörrbaum, A.R., Kochen, L., Langer, J.D., and Schuman, E.M. Local and global influences on protein turnover in neurons and glia. eLife (2018).
DOI Tushev, G., Glock, C., Heumueller, M., Biever, A., Jovanovic, M., and Schuman, E.M. Alternative 3’UTRs modify the localization, regulatory potential, stability, and plasticity of mRNAs in neuronal compartments. Neuron, 98, 495-511. (2018).
DOI Schanzenbaecher, C.T., Langer, J.D., and Schuman, E.M. Time- and polarity-dependent proteomic changes associated with homeostatic scaling at central synapses. eLife 2018;7:e33322. (2018).
DOI Alvarez-Castelao, B., Schanzenbaecher, C.T., Hanus, C., Glock, C., tom Dieck, S., Dörrbaum, A.R., Bartnik, I., Nassim-Assir, B., Ciirdaeva, E., Mueller, A., Dieterich D.C., Tirrell, D.A., Langer, J.D., Schuman, E.M. Cell-type-specific metabolic labeling of nascent proteomes in vivo. Nature Biotechnology, 35: 1196-1201. (2017).
DOI Sambandan, S., Akbalik, G., Kochen, L., Rinne, J., Kahlstatt, J., Glock, C., Tushev, G., Alvarez-Castelao, B., Heckel, A., and Schuman, E.M. Activity-dependent spatially localized miRNA maturation in neuronal dendrites. Science, 355 (6325), 634-637. (2017).
Early Schuman lab contributions and history
Synapses, the points of contact between neurons, can occur 100s of microns from the cell body. The strength of synaptic communication is determined and regulated by proteins that inhabit synapses. Given the distance of synapses from the cell body, how do they get the proteins they need (~550,000 proteins per neuron, per minute) to function? The Schuman lab’s work provided the first evidence that proteins made locally near synapses can be used to enhance synaptic communication, a cellular correlate of memory. This early work and her team’s subsequent studies and technical contributions have had an important impact, creating, expanding and solidifying the field of local translation.
In the first experiments Erin conducted as an Assistant Professor together with her first graduate student, Hyejin Kang, she showed that the growth factor BDNF can induce long-lasting synaptic plasticity, the cellular correlate of memory (Kang & Schuman, Science, 1995). In a paper published just one year later (Kang and Schuman, Science, 1996), they discovered that this plasticity requires local protein synthesis. This observation opened up the field of local translation and showed directly that local translation is important for synaptic function. This paper, together with work from Kelsey Martin (and Eric Kandel), Christine Holt and others, was a driving force for the ensuing exploration of local translation in development and plasticity that has occurred. In the 1980s, Steward and colleagues documented the presence of polyribosomes near synapses in electron micrographs. This important observation didn’t gain as much traction as it deserved in the absence of any functional roles for local translation. Local translation is not only a fundamental cell biological process for the normal function of neurons, it has emerged focal point for many neurodevelopmental disorders including Fragile X, and some autism spectrum disorders.
Over the span of 25 years, the Schuman lab has contributed to the evolving picture of the importance of local translation. For example, in 2001, graduate students Girish Aakalu, Bryan Smith, and colleagues visualized directly the protein synthesis that occurs in dendrites. Using a dendritically targeted GFP-based reporter together with physical isolation of the dendrite, they visualized protein synthesis in living dendrites for the first time. This constituted the first convincing demonstration and visualization of dendritic protein synthesis and the approach has now been used by many labs. The lab went on to demonstrate the critical importance of local translation in many contexts, including its role in synaptic homeostasis (Science, 2004; Cell, 2006, Neuron, 2008) and its role downstream of dopamine receptor signaling (Neuron, 2005). In a more recent study (Neuron, 2012), Schuman’s team used RNA-sequencing to identify over 2500 mRNAs present in dendrites, indicating that local translation of synaptic proteins is much more widespread than previously appreciated. These initial sequencing studies have been complemented by work from many other groups and subsequent work in the Schuman lab (see more recent publications of Tushev, Glock, Perez, Biever, Hafner and Donlin-Asp from the lab).
An important reason for the slow start of the local mRNA translation idea may have been the lack of techniques to address and identify proteins in living cells. Here the Schuman group has also made contributions. In order to study protein translation, one needs to identify and visualize the newly synthesized proteins within cells. Spearheaded by the efforts of postdoctoral fellow Daniela Dieterich, the Schuman team together with her colleague Dave Tirrell (Chemistry, Caltech) invented a method that makes use of artificial amino acids and click chemistry to label newly synthesized proteins. This technology enables scientists to tag, and identify newly synthesized proteins in any cell, tissue or organ (PNAS, 2006). In the following work, (Nature Neurosci., 2010; Nature Methods 2015) the lab developed a variant of this technology that allows one to visualize newly synthesized proteins in situ. In work that was recently published (Nature Biotech, 2017), driven by postdoctoral fellow Beatriz Alvarez-Castelao, the lab has now labelled and identified newly synthesized proteins from individual cell types, giving the entire suite of technologies a genetically encodable and cell-type specific control. This platform can be used to identify proteins that drive cellular changes under normal and diseased states and her team and colleagues have created technology platforms in worm, fly, fish and mouse.




