
Proteomics Core Facility
Dr. Julian Langer
The “Proteomics and membrane mass spectrometry” lab is funded by the MPIs for Biophysics and Brain Research. We specialize in the development of workflows, methods and – in collaboration with instrument vendors – mass spectrometers to address specific questions in structural, molecular and neuro-biology.
Our Methods
See here for impressions
We also engage in external collaborations and are currently a member of SPP 2002 (Small proteins in prokaryotes) by the DFG and the MOEL-SOEL-program by the BMBF.
Recent research highlights
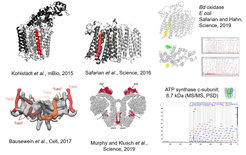
Membrane protein complexes represent challenging targets for bottom-up proteomics. In particular small membrane-embedded subunits often lack proteolytic cleavage sites or do not ionize efficiently in ESI. Together with Bruker Daltonics, we’ve modified a rapifleX MALDI-TOF/TOF mass spectrometer that now allows acquisition of information-rich PSD MS/MS spectra up to m/z 9000. We’ve successfully used this technique to identify multiple new subunits in membrane protein complexes (Kohlstädt et al., mBio, 2015; Safarian et al., Science, 2016; Bausewein et al., Cell, 2017; Murphy et al., Science, 2019; Hahn and Safarian et al., Science, 2019).
HDX-MS of membrane protein complexes.
We successfully characterized ligand binding sites and activity-associated conformational dynamics in multiple membrane protein complexes, including a sodium:proton antiporter (Eisinger et al., PNAS, 2017), a MATE transporter (Eisinger et al., JMB, 2018) and a Molybdate-storage protein (Brünle et al., PNAS, 2019). We currently extend this technique to large membrane protein complexes.
Proteome profiling.
We successfully characterized the proteome remodeling in homeostatic synaptic plasticity in primary hippocampal neurons, making use of the BONCAT technology (Schanzenbächer et al., Neuron, 2016; Schanzenbächer et al., eLife, 2018). We also monitored proteome turnover in this system (Dörrbaum et al., eLife, 2018), and currently investigate its role in homeostatic scaling.
We established a workflow allowing analysis of cell-type specific proteomes from living animals, and investigated proteome dynamics in animals exposed to an enriched environment (Alvarez-Castelao et al., Nature Biotechnology, 2017). A detailed description of the method is available at Alvarez-Castelao et al., Nature Protocols, 2019. Recently, we’ve successfully investigated protein synthesis in neuronal processes (Biever and Glock et al., Science, 2020).
MS Imaging.
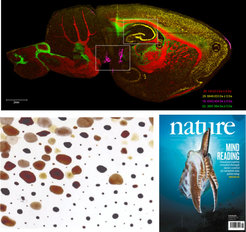
We successfully characterized the molecular components in Sepia chromatophores, that play a key role in their camouflage syste,. Using a combination of UPLC-UV-ESI-MS/MS, MS Imaging and direct infusion UHR-MS, we identified Xanthomatin in these chromatophores, a redox-switchable molecule that had previously been described to play a role in dragonfly maturation and coloration (Reiter et al., Nature, 2018).
Instruments
We operate eight instruments from Bruker, Thermo and Waters, including...
The systems are coupled to nano-UPLCs (Thermo/Dionex Ultimate3000 and easy-1200; Bruker nanoElute) and analytical UPLCs (Thermo/Dionex Ultimate3000; Waters Acquity).

