Solitary ribosomes supply synapses with protein
Scientists reveal surprising insights into protein synthesis in neuron
Learning and memory relies on the ability of our brain cells (“neurons”) to respond adaptively to external inputs. These inputs are integrated at small junctions between neurons called synapses. At synapses proteins the molecules that sense, transmit and store new information. Yet, proteins are unstable- their average life-time is 5-10 days. This raises a big question: how can memories last a lifetime when synaptic proteins are short-lived? Neurons solve this problem in part by directly making proteins “on-site”. At synapses, though, the detection of the conventional protein-making machine (a “polyribosome”- multiple ribosomes associated with a single messenger RNA) has been limited. The apparent limited supply of polyribosomes led to the idea that local protein synthesis is probably only a minor source of synaptic protein. In the upcoming January 31th issue of Science, a team of scientists, led by Erin Schuman, the director of the Max Planck Institute for Brain Research, used in vivo labeling of newly-produced proteins and RNA sequencing of mRNAs engaged with ribosomes in the rodent brain to show that single ribosomes, monosomes, surprisingly represent the active protein-making machine that constitutively supply synapses with proteins.

Memory is encoded and stored in the brain via interactions between neuronal networks. More than 75% of a neuron’s volume consists of long processes, called dendrites and axons, ranging from hundreds of microns to up to 1 meter in length. Through these processes, one neuron can communicate with up to 10000 other neurons at synapses. Synapses are the essential units of memory. These synaptic junctions are “plastic”; their strength can be adapted in response to inputs - a process called “synaptic plasticity”.
Synapses are jam-packed with proteins that have an average lifetime of about 5-10 days. These proteins are the building blocks for the neuron’s essential functions, but also essential for regulating changes in synaptic strength during synaptic plasticity. The big size and complex shape of a neuron imposes huge challenges on supplying synapses with proteins. Over the past 20 years, it has been proposed that neurons solve this problem by shipping out the messenger RNAs (the template for proteins) to dendrites and axons. Surprisingly, however, the long-believed active protein making machines (the polysomes) are infrequent in dendrites - and apparently absent in axons.
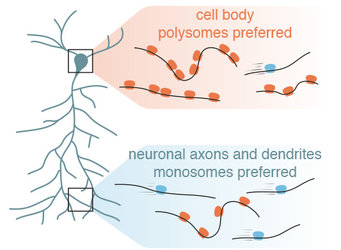
Given the low number/absence of polysomes, it was often assumed that “on-site” protein synthesis had a relatively limited contribution to the synaptic protein levels. “Polysomes can be identified in electron micrographs by a characteristic cluster-like shape”, says Schuman. “In contrast, monosomes cannot be reliably distinguished from other particles using this imaging technique. Therefore, we have very little information about monosomes in the neuron”.
To overcome this limitation, Anne Biever and Caspar Glock, co-first authors of the study, optimized a suite of biochemical tools to analyze the monosome and polysome populations in neuronal cell bodies and dendrites/axons. Polysome profiling, a biochemical fractionation method, was used to determine the number of ribosomes bound to the messages in the different neuronal compartments. “It was surprising to find that mRNAs localized to neuronal processes were more often associated with monosomes”, said Biever.
In mammalian cells, monosomes have been viewed as translationally inactive machines. To gain insights into the translational status of monosomes, the researchers mapped the position of ribosomes on the mRNAs by performing ribosome profiling on isolated monosome and polysome fractions from neuronal compartments. “We were surprised to see that, contrary to the dogma of mammalian cell biology, the monosomes were in the process of active polypeptide elongation in dendrites”, says Glock. “In fact, more than 450 mRNA species were not only translated on monosomes – they showed a preference for monosome-translation. Of these transcripts, many were very important for synaptic function”.
Driven by this finding, the researchers compared the ribosome occupancy between cell bodies and dendrites/axons. “Although most mRNAs showed a similar translational status in the cell bodies and dendrites/axons, our study revealed intriguing examples of transcripts exhibiting differential ribosome occupancy between compartments” says Biever “Among these, we identified key synaptic plasticity proteins”.
The authors then went on to measure protein levels in neuronal processes. “Our results show that the monosome-preferring transcripts often encoded high-abundant proteins. Thus, monosome-translation contributes to maintaining synaptic protein levels”, says Georgi Tushev, head of the institute’s scientific computing group.
“Our finding bridges the gap between the relative shortage of visualized translational machinery in neuronal processes and actual measurements of local translation”, notes Schuman. “The capacity for constitutive “on-site” protein production is much higher than previously assumed, and this, owing to the presence of translationally active monosomes. Monosomes are the solution to the extreme spatial limitations within dendritic spines and axonal boutons, they fit better into the small space and can supply neurons with diverse proteins with limited machinery”, Schuman concludes.
This study was supported by EMBO, the Max Planck Society, the European Research Council and the DFG.
Contact corresponding author: erin.schuman@brain.mpg.de
Publication:
Anne Biever, Caspar Glock, Georgi Tushev, Elena Ciirdaeva, Tamas Dalmay, Julian D. Langer, Erin M. Schuman. Monosomes actively translate synaptic mRNAs in neuronal processes. This paper will be published online by the journal Science on January 30, 2020. DOI: 10.1126/science.aay4991
Links:
https://science.sciencemag.org/cgi/doi/10.1126/science.aay4991